Knowing where you are: custom array indices in Julia
Arrays are a crucial component of any programming language,
particularly for a data-oriented language like Julia. Arrays store
values according to their location: in Julia, given a two-dimensional
array A, the expression A[1,3] returns the value stored at a
location known as (1,3). If, for example, A stores Float64
numbers, the value returned by this expression will be a single
Float64 number.
Julia’s arrays conventionally start numbering their axes with 1,
meaning that the first element of a one-dimensional array a is
a[1]. The choice of 1 vs. 0 seems to generate a certain amount of
discussion. A fairly recent addition to the Julia landscape is the
ability to define arrays that start with an arbitrary index. The
purpose of this blog post is to describe why this might be
interesting. This is really a “user-oriented” blog post, hinting at
some of the ways this new feature can make your life simpler. For
developers who want to write code that supports arrays with arbitrary
indices, see
this documentation page.
Why should we care which indices an array has? A first example
Sometimes arrays are used as simple lists, in which case the indices may not matter to you. But in other cases, arrays are used as a discrete representation of a continuous quantity (e.g., values defined over space or time), and in such cases the array indices correspond to “location” in a way that may be meaningful.
As a simple example, consider the process of rotating an image:
| img | img_rotated |
|---|---|
 |
 |
Many languages provide functions for rotating an image; in Julia, you
can do this with the warp function defined in
ImageTransformations.
Things get a little more “interesting” when you want to compare pixels
in the rotated image to those of the original image. How, exactly, do
these pixels match up? In other words, for a location img[i,j], what
is the corresponding i′,j′ location in img_rotated? In many languages,
figuring out these types of geometric alignments may not be a simple
task; it’s no exaggeration to say that in complex situations (e.g., a
three-dimensional image with a complex spatial deformation) that one
can spend a day or more figuring out exactly how pixels/voxels in two
images are supposed to be compared.
Why is this such a hard problem? The core problem is that, in most cases, the language is essentially “lying” to you about the location of pixels: if arrays always start indexing at 1 along any given axis, the array indices don’t really correspond to an absolute spatial location. An index of 1 means “first index” rather than “spatial location 1.”
So to fix this in Julia, starting with version 0.5 we support arrays with indices that don’t start with 1. Let’s illustrate this by specifying that we want the above rotation to be around a point in the head of the cameraman. Let’s load the image:
julia> using Images, TestImages
julia> img = testimage("cameraman");
julia> summary(img)
"512×512 Array{Gray{N0f8},2}"
summary shows that img is a grayscale image indexed over the
ranges 1:512×1:512. Using any of several approaches (e.g.,
ImageView and paying
attention to the status bar to get the mouse pointer location), we can
learn that the point (y=126, x=251) is in the head of the
cameraman. Consequently, let’s define a rotation around this point:
julia> using Rotations, CoordinateTransformations
julia> tfm = Translation(125,250) ∘ LinearMap(RotMatrix(pi/6)) ∘ Translation(-125,-250)
AffineMap([0.866025 -0.5; 0.5 0.866025], [141.747, -29.0064])
This defines tfm as the composition of a translation (shifting the
head to the origin) followed by a rotation, and then translating back.
(You can get the
composition operator by typing \circ and then hitting TAB.) If we
apply this transformation to the image, we get an interesting result:
julia> img_rotated = warp(img, tfm);
julia> summary(img_rotated)
"-107:592×-160:539 OffsetArray{Gray{N0f8},2}"
Perhaps surprisingly, img_rotated is indexed over the range
-107:592×-160:539, meaning that we access the upper left corner by
img_rotated[-107,-160] and the lower right corner by
img_rotated[592,539]. It’s not hard to see why these numbers arise,
if we see how the corners of img are transformed by tfm:
julia> using StaticArrays
julia> itfm = inv(tfm)
AffineMap([0.866025 0.5; -0.5 0.866025], [-108.253, 95.9936])
julia> itfm(SVector(1,1))
2-element SVector{2,Float64}:
-106.887
96.3597
julia> itfm(SVector(512,1))
2-element SVector{2,Float64}:
335.652
-159.14
julia> itfm(SVector(1,512))
2-element SVector{2,Float64}:
148.613
538.899
julia> itfm(SVector(512,512))
2-element SVector{2,Float64}:
591.152
283.399
This makes it apparent that the output’s indices span the region of the transformed coordinates.
The fact that the output preserves the coordinates makes it trivial to compare the images:
julia> cv = colorview(RGB, paddedviews(0, img, img_rotated, img)...)
paddedviews(0, arrays...) pads input arrays with 0, as needed, to
give them all the same indices, and colorview(RGB, r, g, b) inserts
the grayscale images r, g, and b into the red, green, and blue
channels respectively. If we visualize cv, we see the following:
| around image center | around head (cv) |
|---|---|
 |
 |
The image on the left is for reference, showing what a rotation around the image center would look like when properly aligned. The image on the right corresponds to the steps taken above, and indeed confirms that the rotation is around the head. Alternatively, we can focus on the overlapping portions of these images like this:
julia> inds = map(intersect, indices(img), indices(img_rotated))
(1:512, 1:512)
julia> imgi = img[inds...];
julia> imgri = img_rotated[inds...];
so that colorview(RGB, imgi, imgri, imgi) displays as

Since the indices of the pixels encode absolute spatial location, it’s
trivial to keep track of how different pixels align: pixel i,j in
one image corresponds to pixel i,j in the other. This is true even
if our coordinate transformation were far more complicated than a
simple rotation.
Having motivated why this might be useful, let’s take a step back and walk through array indices a bit more systematically.
A systematic introduction to arrays with arbitrary indices
In Julia, if we define an array
julia> A = collect(reshape(1:30, 5, 6))
5×6 Array{Int64,2}:
1 6 11 16 21 26
2 7 12 17 22 27
3 8 13 18 23 28
4 9 14 19 24 29
5 10 15 20 25 30
then we can refer to a rectangular region like this:
julia> B = A[1:3, 1:4]
3×4 Array{Int64,2}:
1 6 11 16
2 7 12 17
3 8 13 18
For certain applications, one negative to extracting blocks is that there is no record indicating where the new block originated from:
julia> B2 = A[2:4, 1:4]
3×4 Array{Int64,2}:
2 7 12 17
3 8 13 18
4 9 14 19
julia> B2[1,1]
2
So B2[1,1] corresponds to A[2,1], despite the fact that, as
measured by their indices, these are not the same location.
To maintain consistent “naming” of our indices, let’s use the OffsetArrays package:
julia> using OffsetArrays
julia> B3 = OffsetArray(A[2:4, 1:4], 2:4, 1:4) # wrap the snipped-out piece in an OffsetArray
OffsetArrays.OffsetArray{Int64,2,Array{Int64,2}} with indices 2:4×1:4:
2 7 12 17
3 8 13 18
4 9 14 19
julia> B3[3,4]
18
julia> A[3,4]
18
So the indices in B3 match those of A. Indeed, B3 doesn’t even
have an element “named” (1,1):
julia> B3[1,1]
ERROR: BoundsError: attempt to access OffsetArrays.OffsetArray{Int64,2,Array{Int64,2}} with indices 2:4×1:4 at index [1, 1]
Stacktrace:
[1] throw_boundserror(::OffsetArrays.OffsetArray{Int64,2,Array{Int64,2}}, ::Tuple{Int64,Int64}) at ./abstractarray.jl:426
[2] checkbounds at ./abstractarray.jl:355 [inlined]
[3] getindex(::OffsetArrays.OffsetArray{Int64,2,Array{Int64,2}}, ::Int64, ::Int64) at /home/tim/.julia/v0.6/OffsetArrays/src/OffsetArrays.jl:89
In this case we created B3 by explicitly “wrapping” the extracted
array inside a type that allows you to supply custom indices. (You
can retrieve just the extracted portion with parent(B3).) We could
do the same thing by adjusting the indices instead:
julia> using IdentityRanges
julia> ind1, ind2 = IdentityRange(2:4), IdentityRange(1:4)
(IdentityRange(2:4), IdentityRange(1:4))
An IdentityRange is a range with indices that match its values, r[i] == i. (ind1, ind2 = OffsetArray(2:4, 2:4), OffsetArray(1:4, 1:4) would be functionally equivalent.) Let’s use ind1 and ind2 to snip out the region of the array:
julia> B4 = A[ind1, ind2]
OffsetArrays.OffsetArray{Int64,2,Array{Int64,2}} with indices 2:4×1:4:
2 7 12 17
3 8 13 18
4 9 14 19
julia> B4[3,4]
18
This implements a simple rule of composition:
If C = A[ind1, ind2], then C[i, j] == A[ind1[i], ind2[j]]
Consequently, if your indices have their own unconventional indices, they will be propagated forward to the next stage.
This technique can also be used to create a “view”:
julia> V = view(A, ind1, ind2)
SubArray{Int64,2,Array{Int64,2},Tuple{IdentityRanges.IdentityRange{Int64},IdentityRanges.IdentityRange{Int64}},false} with indices 2:4×1:4:
2 7 12 17
3 8 13 18
4 9 14 19
julia> V[3,4]
18
julia> V[1,1]
ERROR: BoundsError: attempt to access SubArray{Int64,2,Array{Int64,2},Tuple{IdentityRanges.IdentityRange{Int64},IdentityRanges.IdentityRange{Int64}},false} with indices 2:4×1:4 at index [1, 1]
Stacktrace:
[1] throw_boundserror(::SubArray{Int64,2,Array{Int64,2},Tuple{IdentityRanges.IdentityRange{Int64},IdentityRanges.IdentityRange{Int64}},false}, ::Tuple{Int64,Int64}) at ./abstractarray.jl:426
[2] checkbounds at ./abstractarray.jl:355 [inlined]
[3] getindex(::SubArray{Int64,2,Array{Int64,2},Tuple{IdentityRanges.IdentityRange{Int64},IdentityRanges.IdentityRange{Int64}},false}, ::Int64, ::Int64) at ./subarray.jl:184
Note that this object is a conventional SubArray (it’s not an
OffsetArray), but because it was passed IdentityRange indices it
preserves the indices of the indices.
A second application: array/image filtering (convolution)
As illustrated above for the image rotation example, a recent release
(v0.6.0) of the Images package put both the power and responsibility
for dealing with arrays with custom indices into the hands of users.
One of the key functions in this package is imfilter which can be
used to smooth or otherwise “filter” arrays. The idea is that starting
from an array A, each local neighborhood is weighted by a “kernel”
kern, producing an output value according to the following formula:
This is the formula for correlation; the formula for another operation, convolution, is very similar.
Let’s start with a trivial example: let’s filter with a “delta
function” kernel, meaning it has value 1 at location 0 and is zero
everywhere else. According to the correlation formula, because kern[J]
is 1 at J==0, this should simply give us a copy of our original array:
julia> using Images
julia> imfilter(1:8, [1])
WARNING: assuming that the origin is at the center of the kernel; to avoid this warning, call `centered(kernel)` or use an OffsetArray
Stacktrace:
[1] depwarn(::String, ::Symbol) at ./deprecated.jl:64
[2] _kernelshift at /home/tim/.julia/v0.6/ImageFiltering/src/imfilter.jl:1049 [inlined]
[3] kernelshift at /home/tim/.julia/v0.6/ImageFiltering/src/imfilter.jl:1046 [inlined]
[4] factorkernel(::Array{Int64,1}) at /home/tim/.julia/v0.6/ImageFiltering/src/imfilter.jl:1016
[5] imfilter at /home/tim/.julia/v0.6/ImageFiltering/src/imfilter.jl:10 [inlined]
[6] imfilter(::UnitRange{Int64}, ::Array{Int64,1}) at /home/tim/.julia/v0.6/ImageFiltering/src/imfilter.jl:5
[7] eval(::Module, ::Any) at ./boot.jl:235
[8] eval_user_input(::Any, ::Base.REPL.REPLBackend) at ./REPL.jl:66
[9] macro expansion at ./REPL.jl:97 [inlined]
[10] (::Base.REPL.##1#2{Base.REPL.REPLBackend})() at ./event.jl:73
while loading no file, in expression starting on line 0
8-element Array{Int64,1}:
1
2
3
4
5
6
7
8
The warning is telling you that Images decided to make a guess about
your intention, that the kernel [1] was intended to be centered around zero. You can
suppress the warning by explicitly passing the following kernel
instead:
julia> kern = centered([1])
OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}} with indices 0:0:
1
julia> kern[0]
1
julia> kern[1]
ERROR: BoundsError: attempt to access OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}} with indices 0:0 at index [1]
Stacktrace:
[1] throw_boundserror(::OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}}, ::Tuple{Int64}) at ./abstractarray.jl:426
[2] checkbounds at ./abstractarray.jl:355 [inlined]
[3] getindex(::OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}}, ::Int64) at /home/tim/.julia/v0.6/OffsetArrays/src/OffsetArrays.jl:94
By using an OffsetArray you have clearly specified your intended indices for kern.
This can be used to shift an image in the following way (by default, imfilter returns its results over the same domain as the input):
julia> kern2 = OffsetArray([1], 2:2) # a delta function centered at 2
OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}} with indices 2:2:
1
julia> imfilter(1:8, kern2, Fill(0)) # pad the edges of the input with 0
8-element Array{Int64,1}:
3
4
5
6
7
8
0
0
julia> kern3 = OffsetArray([1], -5:-5) # a delta function centered at -5
OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}} with indices -5:-5:
1
julia> imfilter(1:8, kern3, Fill(0))
8-element Array{Int64,1}:
0
0
0
0
0
1
2
3
These are all illustrated in the following figure:
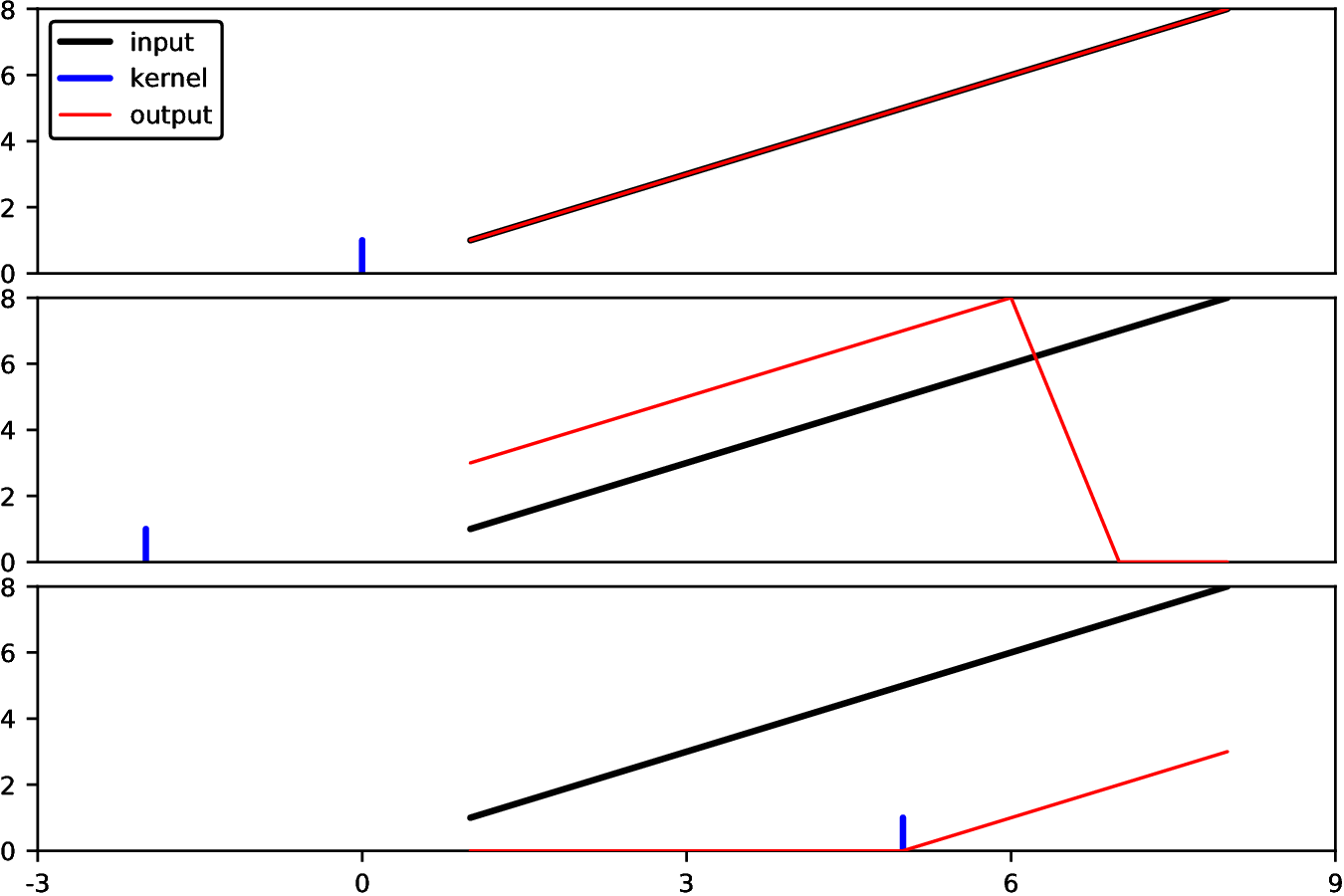
In this figure, we plotted the kernel as if it were at the location corresponding to convolution rather than correlation.
In other programming languages, when filtering with a kernel that has an even number of elements, it can be difficult to remember which of the two middle elements corresponds to the origin. In Julia, that’s not an issue, because you can make that choice for yourself:
julia> kern = OffsetArray([0.5,0.5], 0:1)
OffsetArrays.OffsetArray{Float64,1,Array{Float64,1}} with indices 0:1:
0.5
0.5
julia> imfilter(1:8, kern, Fill(0))
8-element Array{Float64,1}:
1.5
2.5
3.5
4.5
5.5
6.5
7.5
4.0
julia> kern = OffsetArray([0.5,0.5], -1:0)
OffsetArrays.OffsetArray{Float64,1,Array{Float64,1}} with indices -1:0:
0.5
0.5
julia> imfilter(1:8, kern, Fill(0))
8-element Array{Float64,1}:
0.5
1.5
2.5
3.5
4.5
5.5
6.5
7.5
Likewise, sometimes we might have an application where we simply can’t handle the edges properly, and we wish to discard them. For example, consider the following quadratic function:
julia> a = [(i-3)^2 for i = 1:9] # a quadratic function
9-element Array{Int64,1}:
4
1
0
1
4
9
16
25
36
julia> imfilter(a, Kernel.Laplacian((true,)))
9-element Array{Int64,1}:
-3
2
2
2
2
2
2
2
-11
Those weird values on the edges (for which there is no padding that will “extrapolate” the quadratic) might cause problems. Consequently, let’s only extract the values that are well-defined, meaning that all inputs to the correlation formula have explicitly-assigned values:
julia> imfilter(a, Kernel.Laplacian((true,)), Inner())
OffsetArrays.OffsetArray{Int64,1,Array{Int64,1}} with indices 2:8:
2
2
2
2
2
2
2
Notice that in this case, it returned an OffsetArray so that the
values in the result align properly with the original array.
A final application: Fourier transforms
There are many more things you can do with custom indices. As one last illustration, consider the Discrete Fourier Transform, which is defined on a periodic domain. Typically, it’s rather difficult to emulate a periodic domain with arrays, because arrays have finite size. However, it’s possible to define indexing objects which possess periodic behavior. Here we use the FFTViews package, demonstrating the technique on a simple sinusoid:
julia> using FFTViews
julia> a = [sin(2π*x)+0.1 for x in linspace(0,1,16)];
julia> afft = FFTView(fft(a))
FFTViews.FFTView{Complex{Float64},1,Array{Complex{Float64},1}} with indices FFTViews.URange(0,15):
1.6+0.0im
1.498-7.53098im
-0.288537+0.69659im
-0.236488+0.35393im
-0.222614+0.222614im
-0.216932+0.14495im
-0.214217+0.0887316im
-0.212937+0.0423558im
-0.212557+0.0im
-0.212937-0.0423558im
-0.214217-0.0887316im
-0.216932-0.14495im
-0.222614-0.222614im
-0.236488-0.35393im
-0.288537-0.69659im
1.498+7.53098im
Now, as every student of Fourier transforms learns, the 0-frequency bin holds the sum of the values in a:
julia> afft[0]
1.6000000000000003 + 0.0im
Since the mean of a sinusoid is zero, this is (within roundoff error) 16*0.1 = 1.6.
We can also check the amplitude at the Fourier-peak, and explore the periodicity of the result:
julia> afft[1]
1.4980046017247872 - 7.53097769363728im
julia> afft[-1] # negative frequencies are OK
1.4980046017247872 + 7.53097769363728im
julia> afft[64+1] # look Ma, it's periodic!
1.4980046017247872 - 7.53097769363728im
julia> length(indices(afft,1)) # but we still know how big it is
16
Given the periodicity of afft, the commonly-used fftshift function
(e.g., fftshift(fft(a))) can be replaced by afft[-8:7]. While very simple,
these techniques make it surprisingly more pleasant to deal with what
can sometimes become complex and error-prone index gymnastics.
Summary: a user’s perspective
This has only scratched the surface of what’s possible with custom indices. In the opinion of the author, their main advantage is that they can increase the clarity of code by ensuring that “names” (indices) can be endowed with absolute meaning, rather than always being “referenced to whatever the first element of this particular array happens to encode.”
There is quite a lot of code that hasn’t yet properly accounted for the possibility of custom indices—surely, some of it written by the author of this post! So users should be prepared for the possibility that exploiting custom indices will trigger errors in base Julia or in packages. Rather than giving up, users are encouraged to report such errors as issues, as this is the only way that custom indices will, over the course of time, have solid support.
Summary: a developer’s perspective
For some algorithms, there appears to be little reason to ever use arrays with anything but 1-based indices; in such cases, there may be no reason to modify existing code. But if your code has a “spatial” interpretation–where location has meaning–then you just might want to give the new facilities a try.
In transitioning existing code, the author of this post has observed the following tendencies:
-
algorithms that exploit custom indices are sometimes simpler to understand than their “1-locked” counterparts;
-
if you’re porting old code to support custom indices, there’s some bad news: if you had to think carefully about the indexing the first time you wrote it, it usually requires significant investment to re-think the indexing, even if the end result is somewhat simpler.
-
even when a specific algorithm might gain little advantage from supporting arbitrary indices, writing code that is “indices aware” from the beginning is often no harder than writing algorithms that implicitly assume indexing starts at 1.
Developers are referred to Julia’s documentation for further guidance.